Mechanical properties of viral RNA and their relation to function
RNA plays many functional roles in biology beyond simply transmitting genetic information. We are interested in how structure and folding of RNAs interact with cellular processes. We currently focus on RNA structures from viruses whose mechanical properties are important for their function: pseudoknots that stimulate programmed ribosomal frameshifting, and knots that confer resistance to RNases.
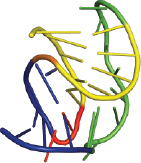
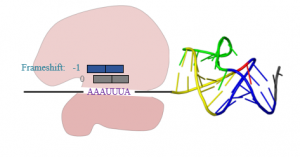
Programmed ribosomal frameshifting (PRF), which allows more than one protein to be made from the same RNA message by causing the ribosome to shift the frame in which it reads the mRNA, occurs frequently in viruses. PRF is induced by specific features in the mRNA: a stimulatory structure like a pseudoknot that the ribosome encounters and has to unfold just as it passes over a “slippery” sequence in the RNA. The mechanism by which stimulatory structures induce PRF remains unclear, but likely involves tension in the mRNA applied by the ribosome as it tries to unfold the stimulatory structure. We have found that efficient stimulatory structures are structurally heterogeneous, and the stimulation of PRF correlates well with the structural heterogeneity. We aim to understand better how PRF is induced by studying the unfolding of stimulatory structures under mechanical tension using optical tweezers, and by observing the motions of single ribosomes as they encounter a stimulatory structure and undergo PRF. We also aim to probe how potential anti-viral drugs that inhibit PRF affect the mechanical properties of stimulatory structures and the dynamics of ribosomes during PRF.
The second focus of our work on RNA involves xrRNAs (exoribonuclease-resistant RNAs), a recently discovered class of non-coding viral RNA that allow viruses to evade key host defense systems when they infect animal and plant cells. xrRNAs resist digestion by host RNases, generating sub-genomic RNA fragments that interact with host proteins to increase viral invasiveness and pathogenicity. They may therefore offer a new target for drugs, which is particularly exciting because they occur in many prevalent but untreatable viruses, including dengue, yellow fever, chikungunya, and West Nile virus. However, many critical questions about xrRNAs remain to be answered, especially regarding the origin of their defining RNase resistance, which is proposed to result from mechanical resistance to digestion of a knot-like topology. We aim to study the mechanical resistance of xrRNAs at the single-molecule level, using optical tweezers to unfold and refold them under tension, thereby clarifying the role of mechanical resistance in xrRNAs, and establishing to what extent it is essential to RNase resistance and how it arises. We also aim to understand how the unusual knotted folds of xrRNAs form, resolving the sequence of steps involved, determining how universal these mechanisms are among different xrRNAs, and probing the role of exoRNase-xrRNA interactions in these mechanisms.

